Metadata
Manage large-scale metadata and tabular data on Pennsieve
To maximally leverage scientific and clinical data, it is critical for researchers to provide detailed and contextual metadata. The Pennsieve platform includes advanced infrastructure to link scientific data to complex metadata schemas. Users can define custom metadata schemas and leverage this to query, explore, and interact with data in context.
Metadata support on the platform is based on the ability to create user-defined data models. Models can be interpreted as the nouns of a dataset (i.e. Subject, Experiment, Study, Diagnosis) and are used to describe the datasets in terms that are meaningful to the user. Each model has properties that describe that model (i.e. properties of a Subject may include Age, Height, Weight, Gender, etc.). In addition, users can define relationships that might exist between records of particular models. For example, a particular Patient X can be enrolled in Study Y. In addition, records can be associated with files allowing users to find raw data associated with specific records.
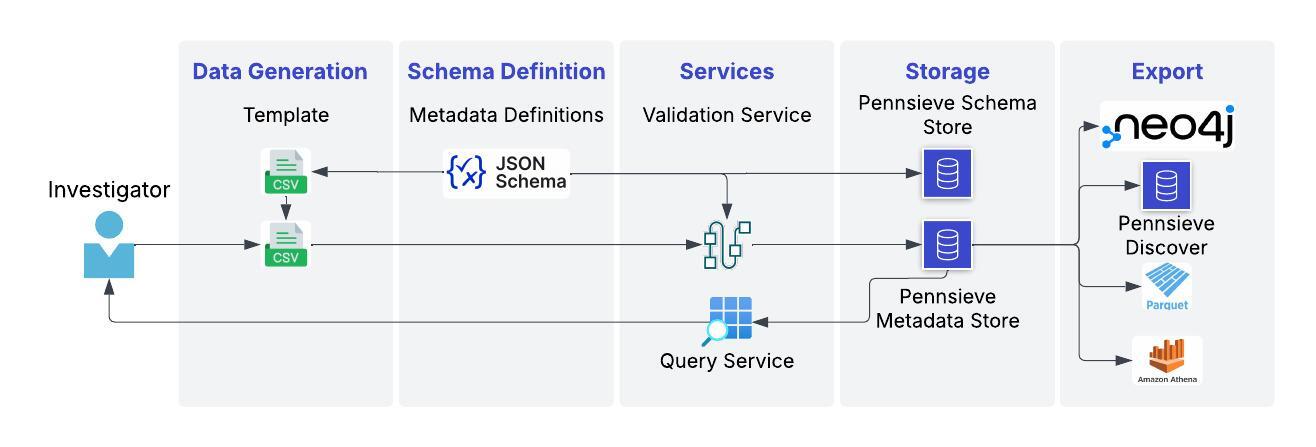
Metadata services are based on JSON and JSON-Schema. They support versioning of both schema and records and is optimized to be AI-Ready. The image below provides a high-level schematic overview of the Pennsieve metadata schema and example processes for ingest and export. 1. Models are defined as JSON Schema objects, 2: Records are defined as JSON objects that are validated against the models during ingest. Metadata records are stored in the Pennsieve Database and are queryable through the platform. 3: Metadata can be exported to several file-formats to enable specific use-cases such as dashboarding, deep graph querying and cohort creation.
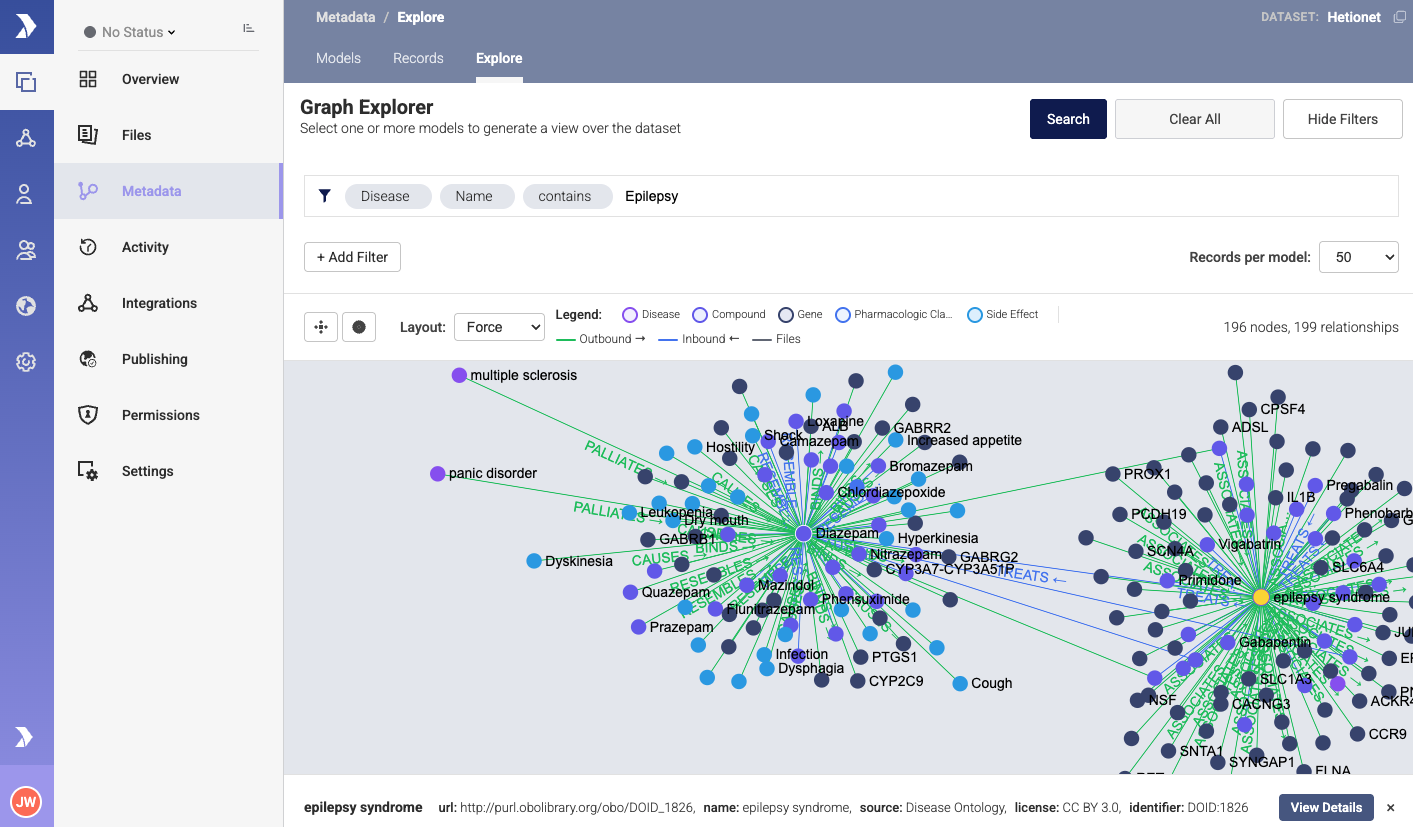
The following screenshot shows an example of the het.io dataset ingested in the Pennsieve platform. Relationships between records allow users to find associations between diseases, drugs, and Genes.
Updated 4 months ago